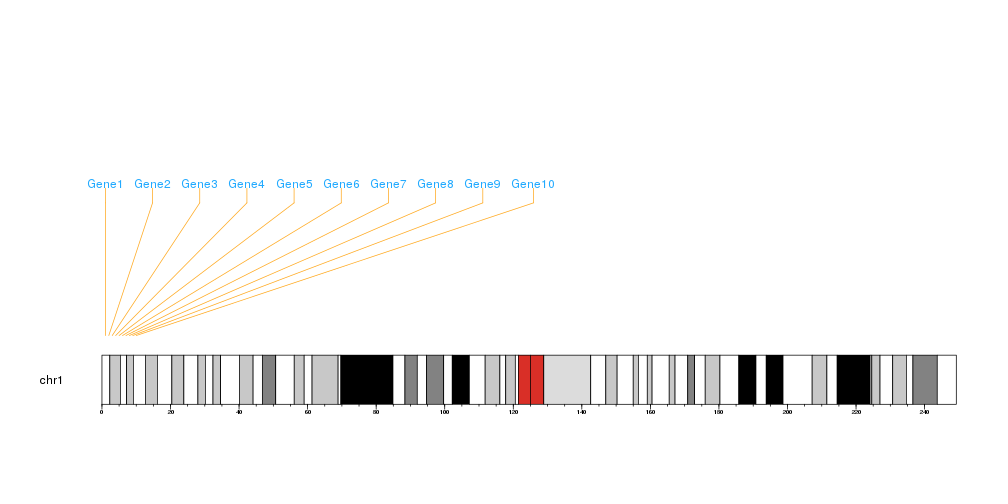
Plotting Genomic Markers
The function kpPlotMarkers can be used to plot markers along the genome, that is, specific positions of the genome with a name: genes, snps, etc…
We will create a set of example markers and plot them on the genome
library(karyoploteR)
markers <- data.frame(chr=rep("chr1", 10), pos=(1:10*10e6), labels=paste0("Gene", 1:10))
kp <- plotKaryotype(chromosomes="chr1")
kpAddBaseNumbers(kp)
kpPlotMarkers(kp, chr=markers$chr, x=markers$pos, labels=markers$labels)
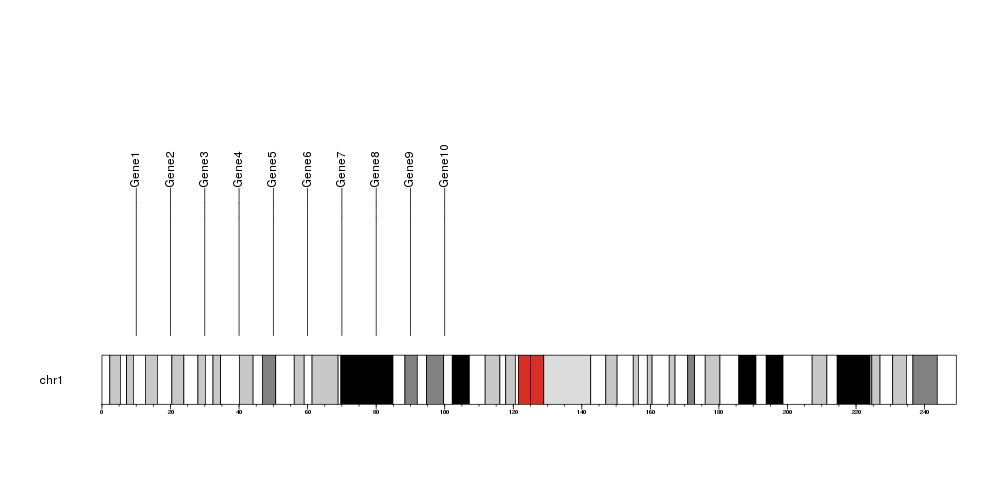
If the positions are closer and the labels will overlap, they will be moved to avoid the overlapping as much as possible. If the label movement is not desired, it can be disabled setting adjust.label.position=FALSE .
markers <- data.frame(chr=rep("chr1", 10), pos=(1:10*1e6), labels=paste0("Gene", 1:10))
kp <- plotKaryotype(chromosomes="chr1")
kpAddBaseNumbers(kp)
kpPlotMarkers(kp, chr=markers$chr, x=markers$pos, labels=markers$labels)
markers2 <- data.frame(chr=rep("chr1", 10), pos=140e6+(1:10*1e6), labels=paste0("OtherGene", 1:10))
kpPlotMarkers(kp, chr=markers2$chr, x=markers2$pos, labels=markers2$labels, adjust.label.position=FALSE)
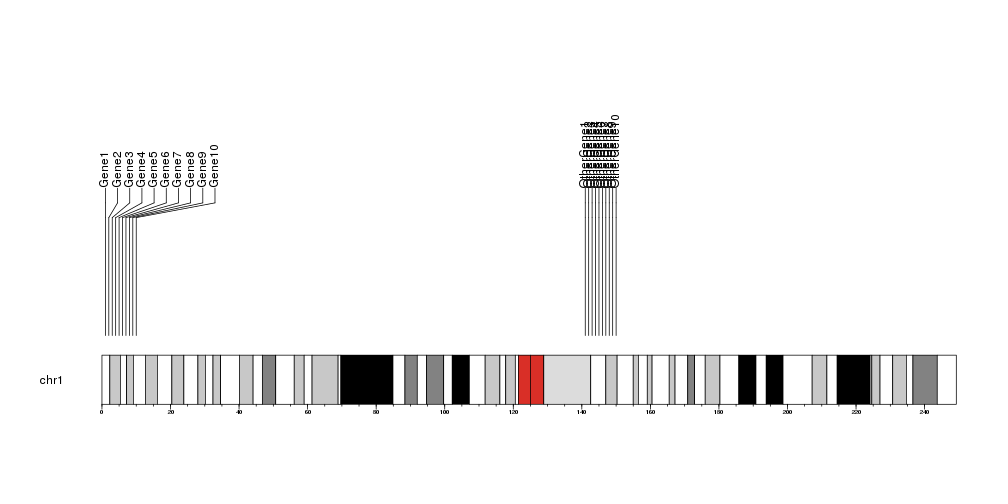
The markers information can be passed as a set of parameters (chr, x, labels) or as a GenomicRanges object with a labels column.
It is possible to customize the marker plotting in several ways. It is possible to specify a different color for the line and the label, to split the line bending in different proportions, to adjust the text orientation and margins and in general to use most of the standard graphics parameters.
markers <- data.frame(chr=rep("chr1", 10), pos=(1:10*1e6), labels=paste0("Gene", 1:10))
kp <- plotKaryotype(chromosomes="chr1")
kpAddBaseNumbers(kp)
kpPlotMarkers(kp, chr=markers$chr, x=markers$pos, labels=markers$labels,
text.orientation = "horizontal", marker.parts = c(0, 0.9, 0.1),
line.color = "#FFAA22", label.color = "#22AAFF",
label.dist = 0.01, max.iter = 1000)